%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import math, warnings
import seaborn as sns
from mpl_toolkits.mplot3d import Axes3D
from random import random
#from mpl_toolkits.mplot3d import axes3d
import matplotlib.pyplot as plt
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import LogisticRegression
from sklearn.linear_model import Lasso
from sklearn.linear_model import Ridge
from sklearn.preprocessing import PolynomialFeatures
from sklearn.model_selection import KFold
sns.set()
#warnings.filterwarnings('ignore')
Logistic regression estimates the probability of an event occurring, such as voted or didn’t vote, based on a given dataset of independent variables. Since the outcome is a probability, the dependent variable is bounded between 0 and 1.The best way to think about logistic regression is that it is a linear regression but for classification problems. Logistic regression essentially uses a logistic function defined below to model a binary output variable (Tolles & Meurer, 2016).The primary difference between linear regression and logistic regression is that logistic regression's range is bounded between 0 and 1. In addition, as opposed to linear regression, logistic regression does not require a linear relationship between inputs and output variables
The odds ratio is the probability to occur some event $A$ divided to the probability of not occur.
$$1) \; \; \; odds(p) = \frac{p(A)}{1-p(A)} $$
where $p(A)\in [0,1]$ Let's see the graphic of the function
p = np.linspace(0,1)
odds = p/(1-p)
plt.plot(p, odds)
plt.xlabel("p(A)", size='20')
plt.ylabel("odds(p)", size='20')
<ipython-input-12-f62918826903>:2: RuntimeWarning: divide by zero encountered in true_divide odds = p/(1-p)
Text(0, 0.5, 'odds(p)')
We can transform the Odds function into another more appropriate for the interpretation, without losing the underlying information. It is called Log-odds or Logit function.
$$2)\; \; \; loggit(P) = \log(Odds)=\log{ \frac{P(A)}{1- P(A)} } $$
plt.plot(p,np.log(odds)) plt.xlabel("p",size='20') plt.ylabel("log-odds",size='20')
The properties of the function log-odds(p) that we should point out :
The logistic regression is based on the assumption that log-odds is a linear dependent on features values of the dataset.
where
$$h(x^i)= \vartheta_0 + \vartheta_1 x^i_{1} + \vartheta_2 x_{2}^i+ ...\vartheta_p x_{p} $$
and $i$ is number of obeservation,
$x^i_m $ feature values,
$\vartheta_0$ intercept,
$\vartheta_{m}$ (weight) slope coeff. for each explanority var.
Let's do some math
If we raise up both sides of eq 3) on a base $e$ :
$$e^{log(\frac{p^i}{1-p^i})} = e^{h(x^i)} \Leftrightarrow $$
$$ \frac{p^i}{1-p^i} = e^{h(x^i)} \Leftrightarrow $$
$$ p^i = e^{h(x^i)}-p.e^{h(x^i)} \Leftrightarrow $$
$$ p^i(1+e^{h(x^i)} ) = e^{h(x^i)} \Leftrightarrow $$
$$p^i= \frac {e^{h(x^i)}} {1 +e^{h(x^i)}} \Leftrightarrow $$
$$p^i= \frac{1}{1 +e^{-h(x^i)} } $$
usualy $p^i$ is written as $\sigma^i$ or $\sigma_i$ and it is called sigmoid
The sigmoid function has very interesting properties. Let's check out for example : $$\sigma =\frac{1}{1 +e^{-(2.x + 4)} }$$
# def sigmoid(x,a,b):
# z = [i*a +b for i in x]
# result = [1/(1+np.exp(-i)) for i in z ]
# return result
def sigmoid(x,a,b):
z = np.array([i*a +b for i in x])
return 1/(1+np.exp(-z))
x = np.linspace(-10,10,30)
y = sigmoid(x,2,4)
plt.plot(x,y)
plt.xlabel("X",size='20')
plt.ylabel(r'$\sigma(X)$',size='20')
Text(0, 0.5, '$\\sigma(X)$')
The graph of sigmoid function is an S-shaped curve as above line in the graph.The main proporties :
def cartesian(start,end,step):
x = y = np.linspace(start,end,step)
return np.transpose([np.tile(x, len(y)), np.repeat(y, len(x))])
def plot(num):
size = np.sqrt(num)
a = cartesian(1, 100, num)
x = np.linspace(-10, 10, 30)
size = int(size)
fig, axs = plt.subplots(size, size,figsize=(25, 25))
for i in range(size):
for j in range(size):
t_1= 2**i/10 + i
t_2 = 3**j - j
axs[i, j].set_title(r'$ \theta_0 = {},\theta_1 = {} $'.format(t_1, t_2), size=20)
y=sigmoid(x, t_1, t_2)
axs[i, j].plot(x, y)
plot(25)
The purpose of Logistic regression is to find that curve that fits as well as possible given dataset.
The logistic regression model is literally a model for the $p$ parameter of a binomial distribution.
$$P(y_i|X,\theta) = \frac{n!}{y_i!(n-1)!} \sigma^{y_i}(1 - \sigma_i )^{n-y_i} $$
when $n=1$
$$P(y_i|x_i,\theta) =\sigma^y_i(1 - \sigma_i )^{1-y_i}$$
Maximum Likelihood Estimation, or MLE for short, is a probabilistic framework for estimating the parameters of a model. In Maximum Likelihood Estimation, we wish to maximize the conditional probability according to observing data $(X)$ given a specific probability distribution and its parameters $(\theta)$, stated formally as:$P(y_i|X,\theta)$
because $y_i$ are independent,we can do joint probability :
$$ argmax_{\Theta} P(y_i|X,\theta) = argmax_{\Theta} P(y_i|x_0,\theta)P(y_i|x_1,\theta)...P(y_i|x_n,\theta)= argmax_{\Theta} \prod_{i=0}^{n}\sigma^y_i(1 - \sigma_i )^{1-y_i}$$
where $\sigma_i$ is sigmoid which our model specific probability distribution function sigmoid.
As same as linear regression we can maximize log Likelihood, because log is monotonic function and if we get minus of log : $$3) \; \; log \big(\prod_{i=0}^{n}\sigma^y_i(1 - \sigma_i )^{1-y_i} \big) = \sum_i^m \big (y^i.\log\sigma^i + (y^i-1)\log{(1 -\sigma^i)}\big)$$
If we get the 3) with minus sign we can instead to maximize we can minimize that function :
$$\Theta = argmax_{\Theta}log \big(\prod_{i=0}^{n}\sigma^y_i(1 - \sigma_i )^{1-y_i} \big) = argmin_{\Theta}-log\big(\prod_{i=0}^{n}\sigma^y_i(1 - \sigma_i )^{1-y_i} \big) $$
Our pupose is to minimize $$4) \; \; \; \mathcal{L} = -\sum_i^m \big (y^i.\log\sigma^i + (y^i-1)\log{(1 -\sigma^i)}\big)$$
that eq is called $ cross-entropy \; loss$ .The next step is to optimized it in order to find its minima.
gradient descent $$5) \; \; \theta_m = \theta_m - \nabla_{\Theta_m} \mathcal{L}(\Theta)*h$$
Let's to find the partial derivate $\frac{\partial \mathcal{L} (\Theta)}{\partial \theta_k}$
$\; \;\frac{\partial \mathcal{L} (\Theta)}{\partial \theta_k} = -\frac{1}{m}\sum_i^m\big(y^i\frac{\partial log(\sigma^i)}{\partial \theta_k}+ (1-y^i)\frac{\partial log(\sigma^i-1)}{\partial \theta_k}\big) $
$= -\frac{1}{m}\sum_i^m\big(y^i \frac{dlog(\sigma^i)}{d\sigma^i(\Theta,x^i_m)}\frac{\partial \sigma^i}{\partial \theta_k}+ (y^i-1) \frac{dlog(1- \sigma^i)}{d\sigma^i(\Theta,x^i_m)}\frac{\partial \sigma^i}{\partial \theta_k}\big) $
$= -\frac{1}{m}\sum_i^m\big(y^i \frac{1}{\sigma^i}\frac{\partial \sigma^i}{\partial \theta_k}+ (y^i-1) \frac{1 }{1- \sigma^i}\frac{-\partial \sigma^i}{\partial \theta_k}\big) $
let's calculate $\frac{\partial \sigma^i}{\partial \theta_k} = \frac{d(\frac{1}{1 +e^{-h(x^i)}})}{dh(x^i)}.\frac{\partial h(x^i)}{\partial \theta_k} =\frac{e^{-h(x^i)}}{(1+e^{-h(x^i)})^2}\frac{\partial h(x^i)}{\partial \theta_k} =\frac{1}{1+e^{- h(x^i)}}(1 -\frac{1}{1+e^{-h(x^i)}} )\frac{\partial h(x^i}{\partial \theta_k} = \sigma^i (1 - \sigma^i\frac{\partial h(x^i)}{\partial \theta_k}) $ and applying it above we achieve
$\; \; \frac{\partial \mathcal{L} (\Theta)}{\partial \theta_k} = -\frac{1}{m}\sum_i^m\big(y^i \frac{1}{\sigma^i}\sigma^i (1 - \sigma^i \frac{\partial h(x^i)}{\partial \theta_k} )+ (y^i-1) \frac{1 }{1- \sigma^i}.-\sigma^i(1 - \sigma^i\frac{\partial h(x^i)}{\partial \theta_k}) $
$= -\frac{1}{m}\sum_i^m\big(y^i\frac{\partial h(x^i)}{\partial \theta_k} -y^i \sigma^i\frac{\partial h(x^i)}{\partial \theta_k} +y^i\sigma^i\frac{\partial h(x^i)}{\partial \theta_k}- \sigma^i\frac{\partial h(x^i)}{\partial \theta_k}\big)$
$= -\frac{1}{m}\sum_i^m\big(y^i\frac{\partial h(x^i)}{\partial \theta_k}- \sigma^i\frac{\partial h(x^i)}{\partial \theta_k}\big)= -\frac{1}{m}\sum_i^m\big(y^i - \sigma^i\big) \frac{\partial h(x^i)}{\partial \theta_k}$
for $\frac{\partial h(x^i)}{\partial \theta_k} = \frac{\partial \theta_mx^i_m}{\partial \theta_k} = \delta_{mk}x^i_m = x^i_m$ applying this result above we will achieve (every k must be replace with m)
For every partial derivate we can write above in matrix form suitable for numy calculation
class BinaryLogisticRegression:
def __init__(self):
self.sample_weight = None
self.X_prime = None
self.Y_prime = None
def fit(self, X, y, C=0.05, I=2500):
self.X_prime, self.Y_prime = self.__validate(X, y)
self.X_prime, self.sample_weight = self.__prepare(self.X_prime, self.Y_prime)
m = 1 / X.shape[0]
for i in range(I):
gradient = m * self.X_prime.T.dot(self.__sigmoid__(self.X_prime, self.sample_weight) - self.Y_prime)
self.sample_weight = self.sample_weight - gradient * C
def score(self, X, y):
X, y = self.__validate(X, y)
y_p = self.predict(X)
res = y - y_p
correct = res[np.nonzero(res == 0)].shape[0]
size = y_p.shape[0]
return correct / size
def predict(self, X):
X, e = self.__validate(X, self.Y_prime)
X, e = self.__prepare(X, self.Y_prime)
return np.round(self.__sigmoid__(X, self.sample_weight))
def __sigmoid__(self, X, wights):
H = X.dot(wights)
return 1 / (1 + np.exp(-H))
def __validate(self, X, y):
X = np.array(X)
# y = np.array(y)
y = np.array(y).reshape(y.shape[0], 1)
return X, y
if False:
raise ValueError("Maximum number of iteration must be positive;"" got (max_iter=%r)" % self.max_iter)
def __prepare(self, X, y):
weights = None
X = np.append(np.ones((X.shape[0], 1)), X, axis=1)
weights = np.ones((X.shape[1], 1))
return X, weights
We will test our implementation on data baknote dataset
data = pd.read_csv('https://archive.ics.uci.edu/ml/machine-learning-databases/00267/data_banknote_authentication.txt')
data.columns = ['variance','skewness','curtosis','entropy','Class']
data
| variance | skewness | curtosis | entropy | Class | |
|---|---|---|---|---|---|
| 0 | 4.54590 | 8.16740 | -2.4586 | -1.46210 | 0 |
| 1 | 3.86600 | -2.63830 | 1.9242 | 0.10645 | 0 |
| 2 | 3.45660 | 9.52280 | -4.0112 | -3.59440 | 0 |
| 3 | 0.32924 | -4.45520 | 4.5718 | -0.98880 | 0 |
| 4 | 4.36840 | 9.67180 | -3.9606 | -3.16250 | 0 |
| ... | ... | ... | ... | ... | ... |
| 1366 | 0.40614 | 1.34920 | -1.4501 | -0.55949 | 1 |
| 1367 | -1.38870 | -4.87730 | 6.4774 | 0.34179 | 1 |
| 1368 | -3.75030 | -13.45860 | 17.5932 | -2.77710 | 1 |
| 1369 | -3.56370 | -8.38270 | 12.3930 | -1.28230 | 1 |
| 1370 | -2.54190 | -0.65804 | 2.6842 | 1.19520 | 1 |
1371 rows × 5 columns
x = data.iloc[:, : -1]
y = data.iloc[:, -1]
scaler = StandardScaler()
x = scaler.fit_transform(x)
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size=0.33, random_state=42)
r = BinaryLogisticRegression()
r.fit(X_train, y_train)
print('predictors weights')
print(r.sample_weight)
print('accuracy')
print(r.score(X_test, y_test))
predictors weights [[-0.79795648] [-3.72638035] [-3.0569379 ] [-2.96198232] [ 0.36577552]] accuracy 0.977924944812362
Let's to test the data on sklearn implemntation
from sklearn.linear_model import LogisticRegression
model = LogisticRegression()
model.fit(X_train, y_train)
LogisticRegression()
print('predictors weights')
print(model.coef_ )
print('accuracy sklearn-learn implementation')
print(model.score(X_test, y_test))
predictors weights [[-4.55543599 -4.47292913 -4.19422475 0.31206293]] accuracy sklearn-learn implementation 0.9801324503311258
The accuracy of sklearn is a little bit better than our implementation.
Similar to the linear regression, even logistic regression is prone to overfitting The Above graph shows that exactly as linear regression logistic model could get overfit by increasing the degree of a polynomial or if the data suffers from multi-colinearity
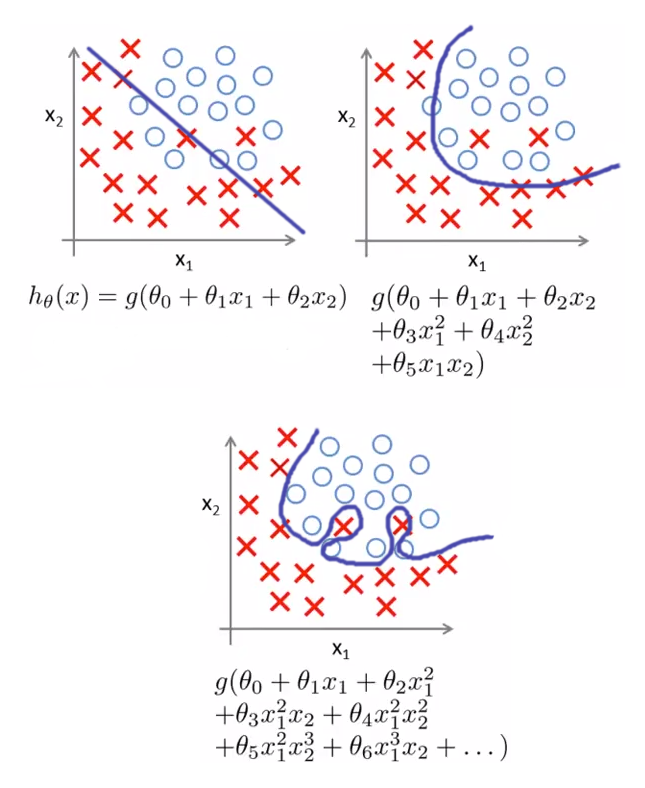
We can use L1, L2 reguralization in the same way as Linear regression to prevent overfitting
L1 Lasso $$ \; \; \mathcal{L} = \sum_i^m \big (y^i.\log\sigma^i + (y^i-1)\log{(1 -\sigma^i)}\big) + \lambda \sum_j^m|\theta_j|$$
Ridge L2 $$ \; \; \mathcal{L} = \sum_i^m \big (y^i.\log\sigma^i + (y^i-1)\log{(1 -\sigma^i)}\big) + \lambda \sum_j^m\theta_j^2 $$